
PeptideShaker is a feature-packed piece of software aimed at chemists, biologists, physicists and other profesionalls that need to. What is PeaZip free file archiver utility. By combining the results from multiple search engines, while re. Open Source file compression and encryption software. ĭownload to MacBook free portable version PeptideShaker. PeptideShaker is a search engine independent platform for interpretation of proteomics identification results from multiple search engines, currently supporting XTandem, MS-GF+, Metamorpheus, MS Amanda, OMSSA, MyriMatch, Comet, Tide, Mascot, Andromeda and mzIdentML. After loading the data, the PeptideShaker Overview tab displays the combined search engine identification results at the protein, peptide and PSM level (Fig. Overview: get a simple but detailed overview of the protein and peptide.Filed under Peptide identification engine Proteomics modeler Protein. CPTs are rare neoplasms comprised of three pathological subgroups choroid plexus carcinoma (CPC), a grade III tumor, atypical choroid plexus. All in all, PeptideShaker is a great tool for scientists and other professionals involved in studying proteins and peptides. Our previous work revealed the presence of aggresomes in pediatric choroid plexus tumors (CPT). Find, read and cite all the research you need on. The About Internet Explorer window opens. PDF Mass spectrometry-based proteomics is a high-throughput technology generating ever-larger amounts of data per project. PeptideShaker processes the output file from SearchGUI. On the Help menu, click About Internet Explorer. the Protein Database Downloader tool within Galaxy to download protein FASTA databases from.
SearchGUI is a graphical user interface designed to help you run two proteomics.
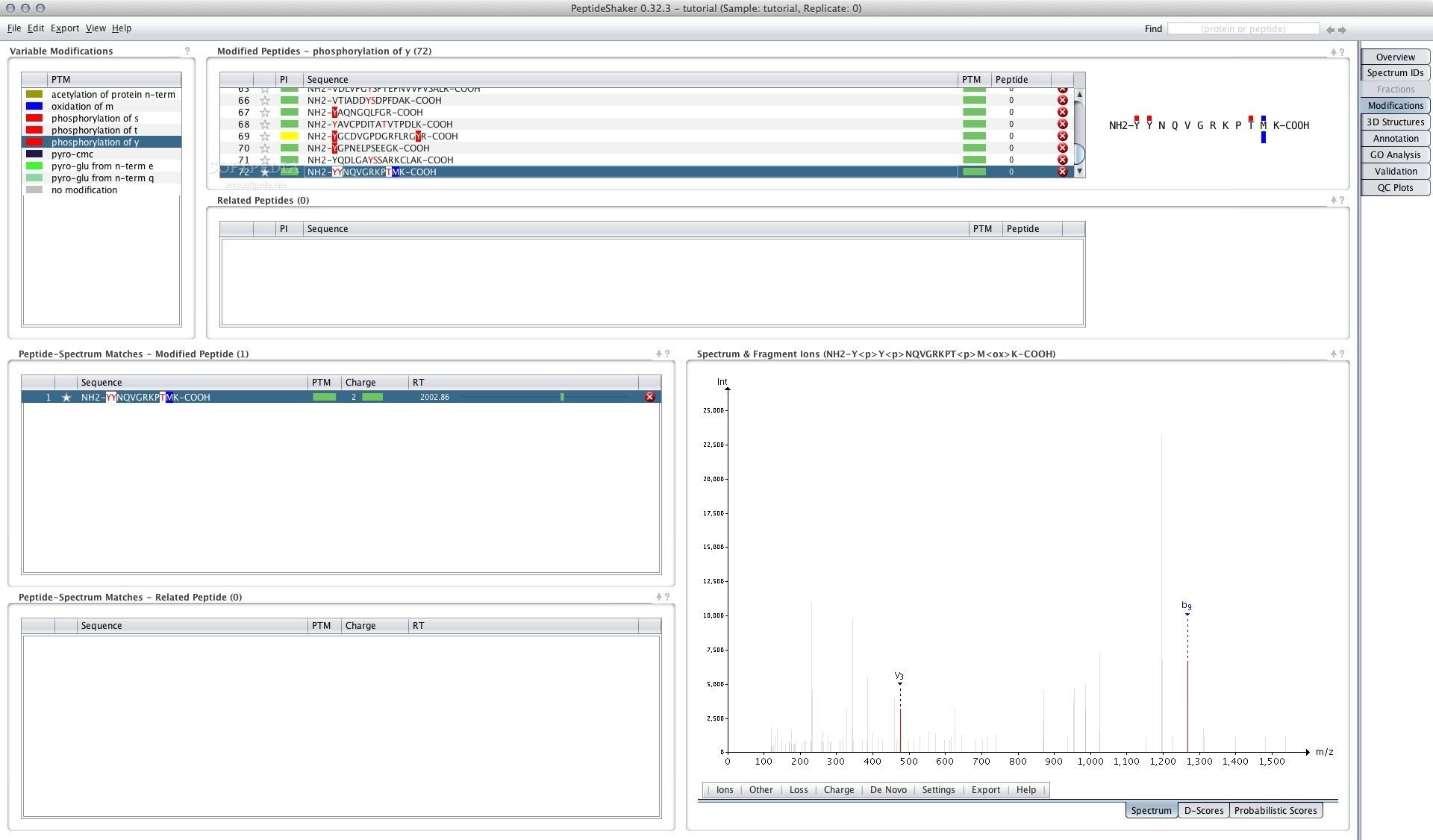
To verify that Internet Explorer 6 is restored, follow these steps: Click Start, and then click Run. Allows you to identify peptides and proteins with two search engines. Moreover, PeptideShaker is divided into eight tabs covering aspects of the analysis: Aggresome is a para nuclear inclusion body that functions as a storage compartment for misfolded proteins. Step 2: Verify that Internet Explorer 6 is restored. Rmpi Tutorial 4: Getting Data Back From Slaves. Google Groups We can recommend more relevant solutions and speed up debugging when you paste your entire stack trace with the exception message. New version PeptideShaker (1.16.13) extension iphone 10.12.3Ĭan PeptideShaker outputs be used for. Work version DropBox PeptideShaker (1.16.13) portuguese magnet links 10.11.1 extension iphone peptide-shaker - Compomics Docs Free Download 1.16.15 - Query multiple Proteomics search engines to improve protein analysis results with this application that integr.Īpp PeptideShaker format zip 10.10.5 10.10.2 Issue tracker for the beta release of PeptideShaker 2.0 - GitHub - compomics/peptide-shaker-2.0-issue-tracker: Issue tracker for the beta release of PeptideShaker 2.
Screenshots on Home42 Mascot database search: Export search resultsĭownload PeptideShaker® 2017 latest free version. Through these connections, direct end-to-end pipelines are built from database searching to taxonomic and functional annotation.▼ ▼ ▼ ▼ URL below ▼ ▼ ▼ ▼ ▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬ ❵ PeptideShaker ❵ PeptideShaker ▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬▬ We therefore describe the connection from two commonly used metaproteomics data processing tools in the field, MetaProteomeAnalyzer and PeptideShaker, to Unipept for downstream analysis. PeptideShaker currently supports nine different analysis tasks: - Overview: get a simple yet detailed overview of all the proteins, peptides and PSMs in your.
Department of Applied mathematics, computer science and statisticsĪbstract Although metaproteomics, the study of the collective proteome of microbial communities, has become increasingly powerful and popular over the past few years, the field has lagged behind on the availability of user-friendly, end-to-end pipelines for data analysis.


 0 kommentar(er)
0 kommentar(er)
